The Power of Iso-Seq in Gene Discovery and Annotation
Discover how Iso-Seq technology revolutionizes gene discovery and annotation, enabling precise identification of novel genes, isoforms, and non-coding RNAs. Learn more!

Gene discovery and annotation serve as essential cornerstones in our ongoing endeavor to decipher the intricate mysteries of biological processes, disease mechanisms, and evolutionary connections. They provide the foundational knowledge needed to advance our understanding of life at its most fundamental level. Over the years, conventional RNA-Seq has played a pivotal role in gene expression analysis, profoundly transforming our comprehension of transcriptome dynamics and shedding new light on the intricate web of genetic interactions. Despite its significant contributions, conventional RNA-Seq's reliance on short readings has inherently constrained its ability to fully capture the complex and nuanced structures of transcripts. This limitation has spurred the development of more advanced sequencing technologies, and among these emerging innovations, Iso-Seq stands out as a trailblazer, pushing the boundaries of what we can achieve in gene discovery and annotation.
Leveraging PacBio's Single-Molecule Real-Time (SMRT) sequencing platform, Iso-Seq offers a groundbreaking approach to gene discovery and annotation by providing a full-length view of RNA transcripts. This technology not only enhances the precision of gene annotations but also facilitates the identification of novel genes, isoforms, and non-coding RNAs, thereby pushing the boundaries of genomics research.
Overview of Iso-Seq Technology
Iso-Seq, or Isoform Sequencing, represents a paradigm shift in transcriptome analysis. By harnessing the power of PacBio's SMRT sequencing, Iso-Seq directly captures full-length cDNA sequences, encompassing both untranslated regions (UTRs) and polyadenylated tails. In stark contrast to short-read RNA-Seq, which fragments and assembles RNA molecules, Iso-Seq sequences entire transcripts without the need for assembly. This method yields high-fidelity reads spanning up to 10 kb or more, ensuring the preservation of long, complex transcripts' integrity.
The capability of Iso-Seq to produce high-quality, full-length transcripts underscores its invaluable significance in studying gene isoforms, alternative splicing events, and fusion genes. These features, often overlooked by traditional RNA-Seq, contribute to the transcriptome's diversity and complexity. Consequently, Iso-Seq has found widespread application in genomic research, encompassing genome-wide annotation, isoform discovery, and single-cell analysis. Researchers now possess an unprecedented tool to uncover the transcriptome's hidden facets, previously obscured by the limitations of short-read sequencing.
Importance of Gene Discovery and Annotation in Genomics
Accurate gene discovery and annotation are indispensable for deciphering the functional elements within genomes. RNA-Seq, despite its robustness in gene expression analysis, struggles to offer a comprehensive view of complex transcript structures due to its short-read nature. Iso-Seq rises to meet this challenge, presenting a complete, uninterrupted view of full-length transcripts. This holistic approach enhances the reliability and precision of gene annotations, empowering researchers to identify previously unannotated genes and isoforms.
In studies of human and rat brains, Iso-Seq has revealed numerous novel transcripts undetected by short-read RNA-Seq, thereby refining gene annotations. Similarly, in agricultural genomics, Iso-Seq has uncovered thousands of novel isoforms and genes, vastly expanding our understanding of plant and animal transcriptomes. This technology's transformative impact on gene annotation is poised to revolutionize genomics research, providing deeper insights into gene function, regulation, and disease mechanisms.
Gene Discovery with Iso-Seq
Iso-Seq has emerged as a transformative tool in gene discovery, empowering researchers to pinpoint novel genes and transcripts with exceptional accuracy. The technology's ability to sequence full-length RNA molecules without prior assembly steps grants a more extensive view of gene structures, uncovering previously concealed isoforms and rare transcripts.
Iso-Seq's direct sequencing of full-length RNA captures the entire transcript, including the 5' and 3' ends, eliminating the need for assembly. This facilitates the identification of previously uncharacterized isoforms, including those that are rare or low in abundance. Notably, Iso-Seq excels in detecting alternative splicing events, such as splice-site variations, intron retention, and exon skipping, providing granular information on splice junctions. This level of detail is unattainable using traditional short-read sequencing technologies, rendering Iso-Seq indispensable in the discovery of novel isoforms.
Furthermore, Iso-Seq can uncover previously unidentified genes, even in the absence of a reference genome. By generating full-length transcript data, Iso-Seq facilitates gene model reconstruction and the identification of novel genes overlooked by conventional annotation methods. The ability to capture both coding and non-coding RNA molecules, such as miRNAs and lncRNAs, also allows for the detection of these crucial regulatory elements pivotal in gene expression and disease regulation.
Case Studies of Gene Discovery Using Iso-Seq
Iso-Seq's impact on gene discovery is evident through its diverse applications in both plant and animal genome research. In plant genomics, Iso-Seq has refined gene models and augmented genome annotation accuracy. For instance, in species like wheat and maize, Iso-Seq has provided valuable insights into gene structures, splice junctions, and alternative splicing events. These revelations have improved the annotation of complex plant genomes and enhanced our understanding of gene regulation under various environmental conditions.
In animal genomics, particularly in primates, Iso-Seq has enabled the discovery of new transcripts and isoforms, contributing to the comprehension of evolutionary relationships. By unveiling transcript variations previously undetectable, Iso-Seq has broadened our knowledge of species-specific gene evolution and functional diversity. Additionally, Iso-Seq has demonstrated its prowess in identifying non-coding RNAs often neglected in traditional sequencing approaches. In cancer research, for example, Iso-Seq has uncovered disease-associated fusion genes and non-coding RNAs that could serve as potential biomarkers for diagnosis and therapy, paving new pathways for targeted treatments.
Gene Annotation and Iso-Seq
Integrating Iso-Seq data into gene annotation pipelines has been shown to improve the accuracy and completeness of gene models by generating full-length transcripts. This technology enables researchers to refine gene annotations, presenting gene structure and isozyme diversity more accurately. Iso-Seq is particularly effective in refining existing gene models by correcting misannotated splice sites, extending gene boundaries, and filling gaps in the reference genome. In crop species such as wheat and maize, Iso-Seq has been instrumental in enhancing reference genomes, improving the annotation of complex gene families, and providing more accurate characterization of gene structure. These improvements have informed functional genomics studies and provided more reliable explanations of gene regulation.
Additionally, Iso-Seq data are indispensable for capturing the full complexity of gene isozymes, especially in cases where alternative splicing events result in diverse transcript variants. This approach enables precise annotation of isozyme structures, leading to a more comprehensive understanding of gene expression and regulation. Iso-Seq has been shown to provide enhanced accuracy and completeness in gene annotation compared to traditional RNA-Seq methods. Short-read sequencing has been observed to have limitations in accurately identifying complex splicing events and adequately resolving long repetitive regions.
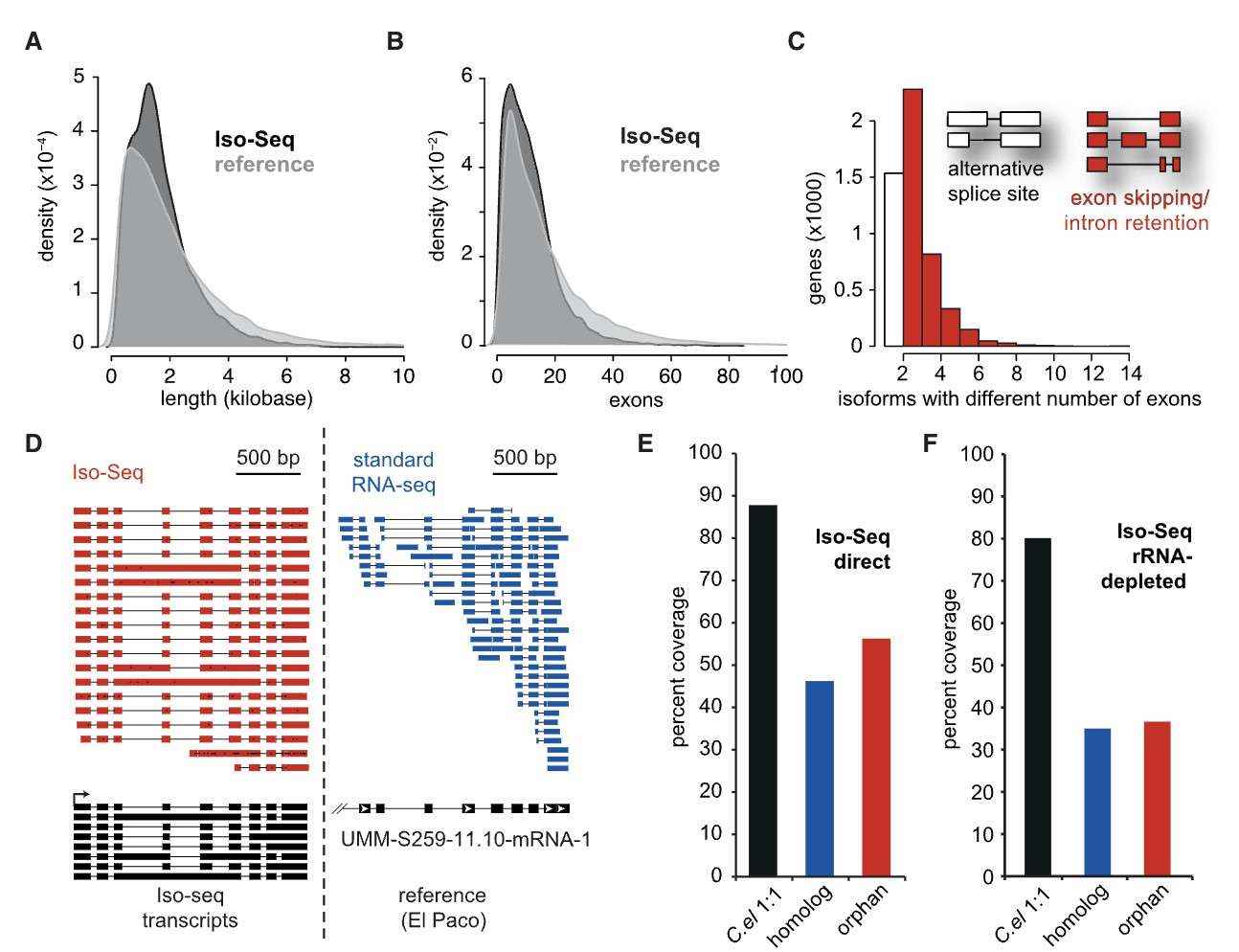
Iso-Seq improves gene annotation, identifies alternative splicing, and can distinguish different evolutionary gene classes by gene structure (Werner et al., 2018)
In contrast, Iso-Seq has been demonstrated to directly sequence full-length transcripts, ensuring comprehensive capture of all splice variants and isoforms. This approach eliminates the need for computationally intensive transcript assembly, reducing the probability of false positives and enhancing the precision of genome annotation.
In addition to enhancing the accuracy of gene annotation, Iso-Seq provides vital data that can propel functional genomics studies forward. By capturing the full spectrum of gene isoforms, Iso-Seq enables a more comprehensive analysis of gene regulation, expression patterns, and functional diversity. This opens the door to novel discoveries in gene function and disease mechanisms.
Applications of Iso-Seq in Genomics and Transcriptomics
Iso-Seq's utility transcends mere gene annotation and discovery, carrying profound implications for functional and comparative genomics. By offering comprehensive insights into gene expression patterns, isoform variety, and alternative splicing mechanisms, Iso-Seq has emerged as a powerful instrument in unraveling the complexities of gene regulation and evolutionary dynamics.
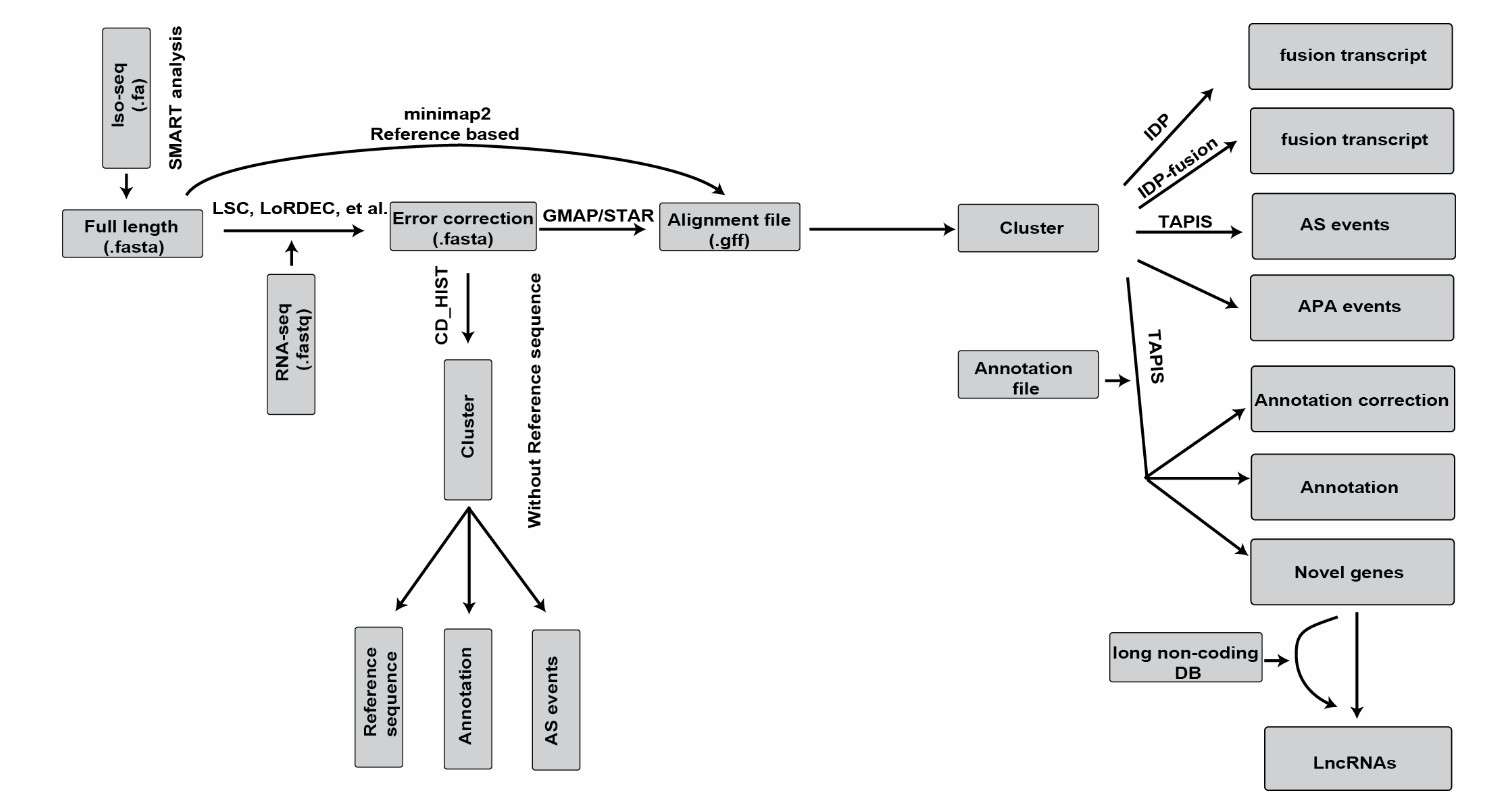
The scheme of the bioinformatics tool and its biological application for Iso-Seq (Gao et al., 2019)
In the realm of functional genomics, Iso-Seq plays a pivotal role by empowering researchers to gain deeper understanding of gene regulation and expression profiles. By scrutinizing isoform diversity across various tissues or disease conditions, Iso-Seq can elucidate how specific gene variations contribute to physiological functions or pathological processes. These revelations are immensely beneficial in comprehending gene functionalities and pinpointing potential therapeutic targets for interventions.
Moreover, Iso-Seq's capacity to detect alternative splicing events enables researchers to explore the impact of these variations on gene functions, including the generation of diverse protein isoforms with unique biological activities. This is particularly significant in studying intricate traits and diseases, such as cancer, where alternative splicing frequently plays a central role in disease progression.
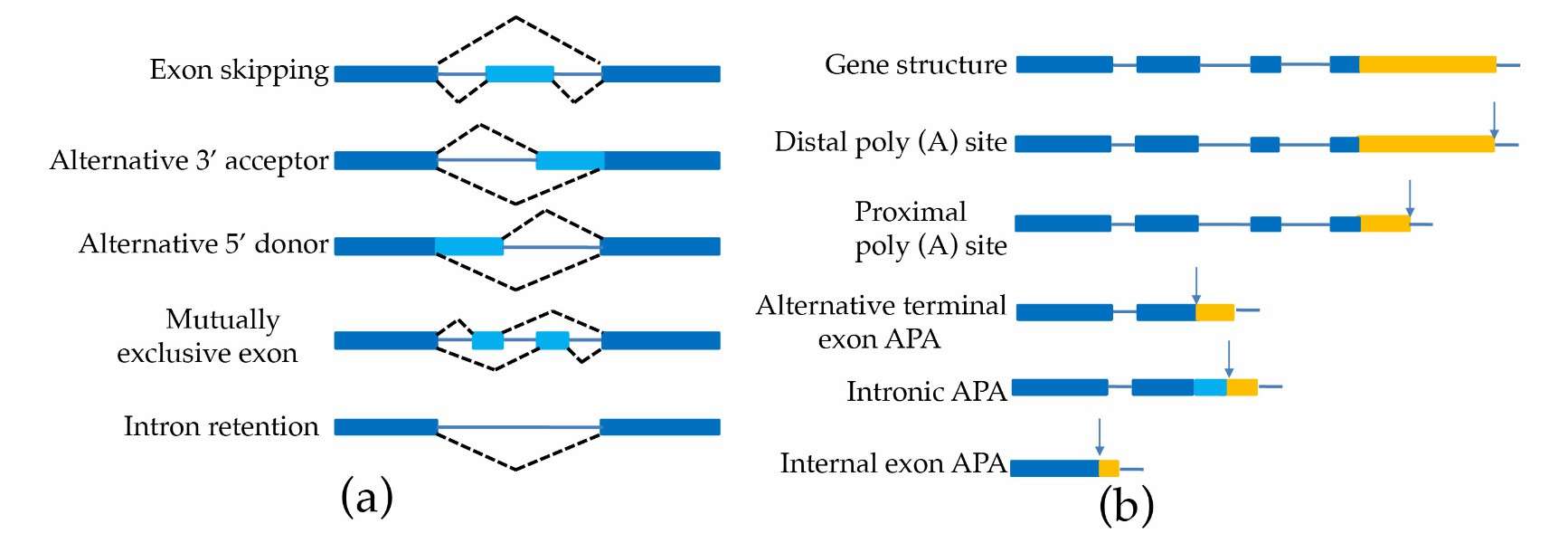
Schematic representation of alternative splicing and alternative polyadenylation (Pacholewska et al., 2024)
The high-resolution data provided by Iso-Seq is also invaluable in comparative genomics studies. By comparing isoform diversity and gene structures among species, researchers can uncover evolutionary discrepancies in gene regulation and function. This has been demonstrated in studies examining gene evolution in both plants and animals, revealing the expansion or contraction of certain gene families over time and the role of alternative splicing in species-specific traits.
Cross-species comparisons of gene isoforms can further offer insights into the evolutionary origins of complex traits and facilitate the identification of conserved regulatory mechanisms. This approach holds promise in shedding light on the genetic foundation of critical biological processes, such as development, immunity, and adaptation to environmental stressors.
Conclusion
Iso-Seq has undoubtedly revolutionized gene discovery and annotation, offering unprecedented insights into gene structures, isoform diversity, and non-coding RNAs. By capturing full-length RNA sequences, Iso-Seq has significantly enhanced the accuracy and completeness of genome annotations, leading to a more profound understanding of gene regulation and expression.
Iso-Seq's remarkable capacity to generate comprehensive transcript data has significantly enhanced gene annotations, facilitating a more precise depiction of gene structures and isoform diversity. This advancement has had a transformative effect on functional genomics, offering novel insights into gene function, disease mechanisms, and evolutionary relationships.
As sequencing technologies continue to advance, Iso-Seq is poised to further elevate its accuracy and efficiency. Future research will focus on integrating multi-omics data, encompassing transcriptomics, proteomics, and metabolomics, to provide a holistic understanding of biological systems. This integrative approach will not only refine our comprehension of gene function but also accelerate the development of personalized medicine and precision agriculture. Iso-Seq's leadership in this field is poised to usher in a new era of breakthroughs, paving the way for a more comprehensive understanding of transcriptomes across diverse organisms. As we move toward a more comprehensive view of gene expression, splicing, and regulation, Iso-Seq's ability to capture and annotate previously overlooked elements will continue to play a critical role in both basic and applied biological research.
The increasing accessibility and affordability of Iso-Seq technology, when combined with its integration into standard genomics workflows, will expand its applications in areas such as functional genomics, disease biomarker discovery, and evolutionary studies. Iso-Seq's ability to identify novel isoforms and non-coding RNAs holds promise for revealing new layers of genetic regulation, with the potential to transform our approach to treating diseases like cancer, neurodegenerative disorders, and autoimmune conditions.
As we continue to refine our understanding of transcriptomic diversity and gene regulation, Iso-Seq will undoubtedly remain at the forefront of genomics, guiding researchers in their quest to unravel the complex molecular networks that govern cellular processes. Its contributions will help address some of the most pressing questions in biology and medicine, ultimately leading to more effective therapeutic strategies and a deeper understanding of the underlying mechanisms of health and disease.
In conclusion, Iso-Seq is more than just a sequencing method—it is a powerful tool that will continue to shape the future of gene discovery, annotation, and functional genomics. By providing unparalleled insights into the full complexity of the transcriptome, Iso-Seq is driving a new era of genomics research that promises to transform our understanding of life at the molecular level.
References:
- An, Dong et al. "Isoform Sequencing and State-of-Art Applications for Unravelling Complexity of Plant Transcriptomes." Genes 9 (2018). doi:10.3390/genes9010043
What's Your Reaction?


















